| Tutorial - Advanced Interpretation of Volumetric Data |
|
Sculptor can detect alpha-helices and other filaments in 3D EM maps (Rusu and Wriggers, 2012, Rusu et al., 2012). Maps can first be denoised using our DPSV filter (Starosolski et al., 2012). 1. Work flow for denoising maps using DPSVHere is the HIV virion example from our manuscripts (Starosolski et al., 2012, supplemtary data 1 in Rusu et al., 2012):
2. Work flow for detecting alpha helices in intermediate resolution mapsThe following video (shown at half size) descibes the necessary steps in tracing alpha helices. You can download the video here for full size viewing. You can also download the map file emd_1740_AI.situs and the PDB file 3C92_AI.pdb to follow the video along. Most of the VolTrac parameters shown in the video are intuitive, but for sake of clarity we illustrate here the geometry of the template used in the tracing: 3C92_AI.pdb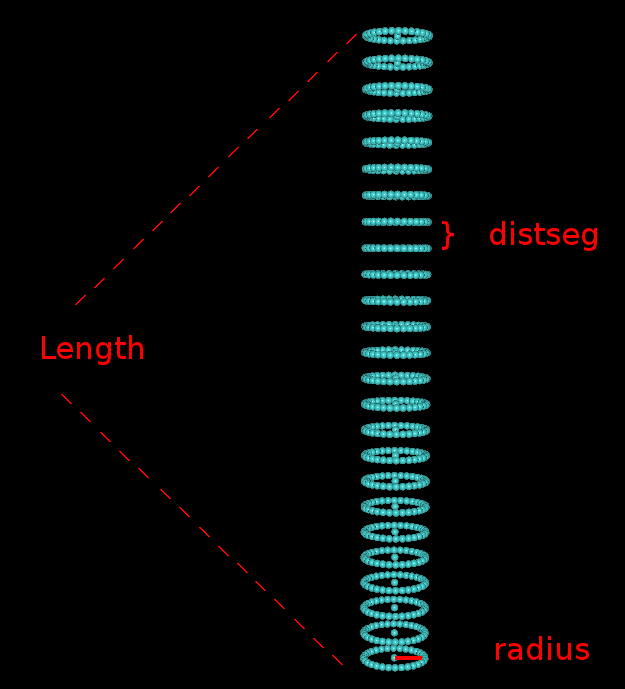 |
| 1. Load PDB |  |
| 2. Show Tubes |  |
| 3. Increase Radius |  |
| 4. Output | 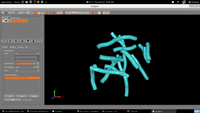 |
Often, a user may wish to manually edit the traces to eliminate false positives (e.g. alpha helices placed in beta sheet regions). The manual editing and merging of the results is described in the following series of screenshots (click on the thumbnails to see the full size figures):
| 5. Show Tubes | 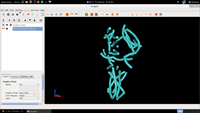 |
| 6. Select Extract | 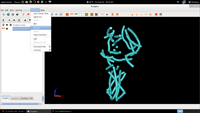 |
| 7. Extract Tubes | 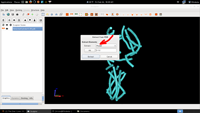 |
| 8. Hide Results |  |
| 9. Change Rendering |  |
| 10. Merge Highlighted |  |
4. Work flow for tracing filaments in tomograms
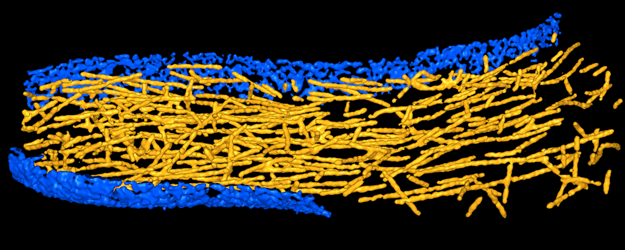
If you are
interested in tracing filamentous density in 3D maps of
cellular structures from tomography or other in vivo imaging
techniques, we recommend to follow first the tutorials in steps 2 and 3
above to learn the basic functionality of the VolTrac algorithm.
Then, refer to Rusu
et al., 2012 which describes in the
main text the parameters and steps taken in the tracing of actin
filaments in a filopodium protrusion. Supplementary data files 2-6 of
this paper comprise snapshots of this workflow. Supplementary
data file 4 lists the steps taken prior to VolTrac, whereas
the VolTrac
parameters are given in the main text of the paper. The final results
shown in Fig. 3B of the paper are provided in supplementary file 6 for
comparison purposes. You can visualize these traces as described for
alpha-helices under step 3 above.
